R programming for beginners (GV900)
Lesson 15: ANOVA
Friday, January 19, 2024
Video of Lesson 15
1 Outline
One way ANOVA
Two way ANOVA
Repeated measures ANOVA
2 Basic concepts
- ANOVA vs t-test
t-test is used to compare two groups, which could be two dependent samples or two independent samples.
ANOVA is used to compare three or more groups, which could be three or more dependent samples or three or more independent samples.
ANOVA: ANalysis Of VAriance
ANOVA is a statistical method used for comparing means among three or more groups to determine if there are statistically significant differences between them.
ANOVA works by partitioning the total variance observed in a dataset into different components: the variance due to differences between groups and the variance due to differences within groups.
ANOVA produces an F-statistic, which compares the variance between group means to the variance within the groups. If the calculated F-value is greater than the critical value from an F-distribution table, it suggests that there are significant differences between at least two of the group means.
3 One-Way ANOVA
One-way ANOVA is used to compare the means of three or more independent (unrelated) groups to determine if at least one of the groups differs significantly from the others.
In one-way ANOVA, we need to find f-value:
F=VaraVarb=s2as2b=MSBMSE
- To show how to calculate f-value, we use the follow simple blood presure data set:
young middle old
1 121 126 131
2 123 128 133
3 125 130 135
4 127 132 137- First, we calculate the mean of each group:
young mean_young middle mean_middle old mean_old
1 121 124 126 129 131 134
2 123 124 128 129 133 134
3 125 124 130 129 135 134
4 127 124 132 129 137 134- Second, we calculate the Sum of Squares of Error (SSE) and Sum of Squares Between Groups (SSB):
SSE=k∑i=1ni∑j=1(xi,j−ˉxi)2
SSB=k∑i=1ni(ˉxi−ˉˉx)2
SSE
1 60Code
total_mean <- mean(c(bp$mean_young, bp$mean_middle, bp$mean_old))
SSB <- bp |>
distinct(mean_young, mean_middle, mean_old) |>
mutate(SSB_young = 4 * (mean_young - total_mean)^2,
SSB_middle = 4 * (mean_middle - total_mean)^2,
SSB_old = 4 * (mean_old - total_mean)^2) |>
summarise(SSB = sum(SSB_young) + sum(SSB_middle) + sum(SSB_old))
SSB SSB
1 200- Now, let’s take a look at the degrees of freedom of the SSE (dfe) and SSB (dfb):
dfe=N−k=12−3=9
dfb=k−1=3−1=2
- Then, let’s calculate the Mean Square Between Groups (MSB) and Mean Square of Error (MSE):
MSB=SSBdfb
MSE=SSEdfe
SSB
1 100 SSE
1 6.666667- Finally, we can calculate the f-value:
F=MSBMSE
| Source | Sum of S quares | Degrees of Freedom | Mean Square | F-value |
|---|---|---|---|---|
| Between Groups | SSB=k∑i=1ni(ˉxi−ˉˉx)2 | dfb=N−k | MSB=SSBdfb | F=MSBMSE |
| Within Groups | SSE=k∑i=1ni∑j=1(xi,j−ˉxi)2 | dfe=k−1 | MSE=SSEdfe | |
| Total | SST=SSE+SSB | dftotal=dfe+dfb |
- Fortunately, we don’t have to calculate the f-value manually. We can use R to do the test:
Df Sum Sq Mean Sq F value Pr(>F)
group 2 200 100.00 15 0.00136 **
Residuals 9 60 6.67
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1In this example, the p-value is 0.00136, which is less than 0.05. Therefore, we reject the null hypothesis and conclude that there is a significant difference in blood pressure among the three age groups.
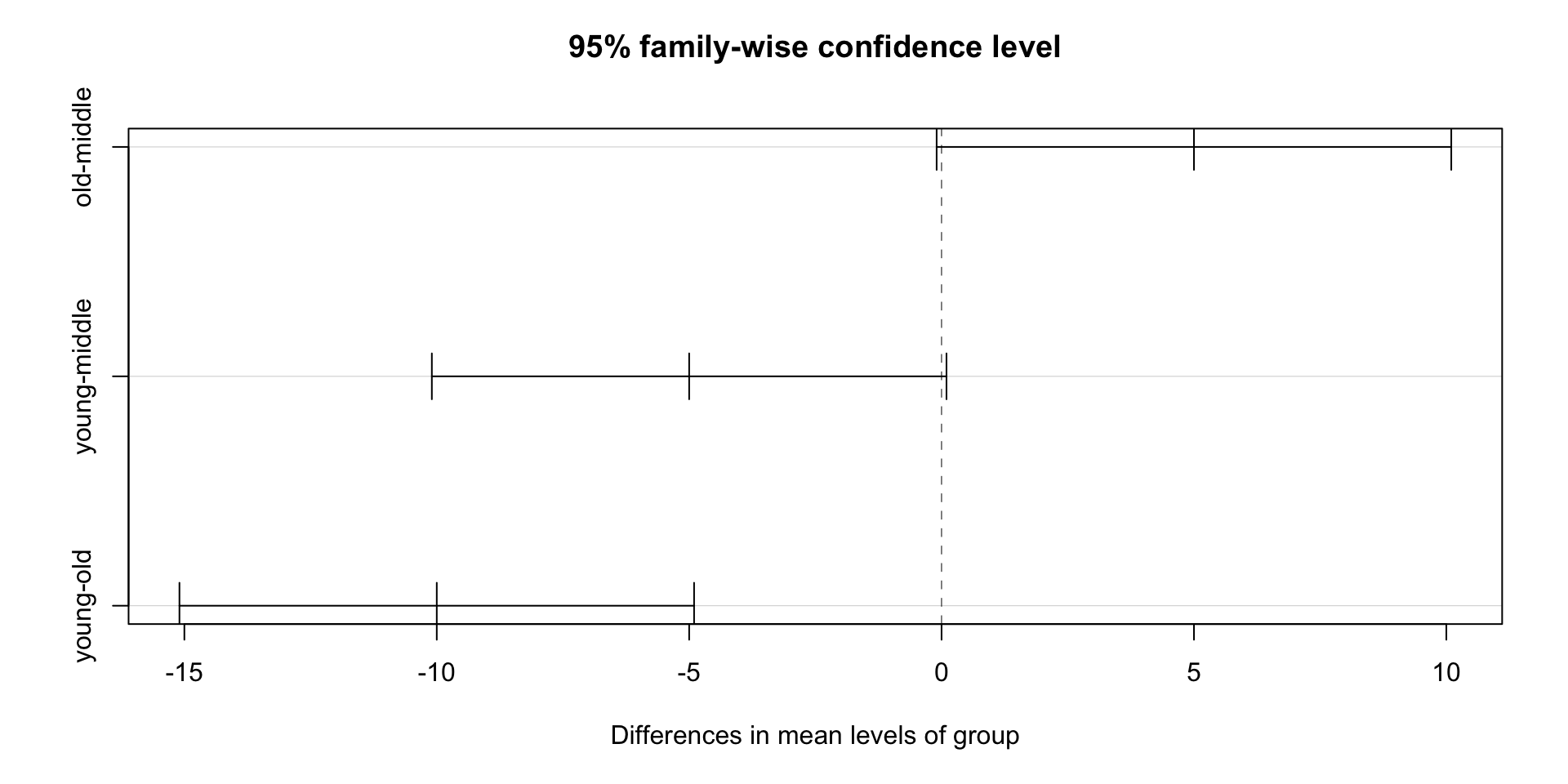
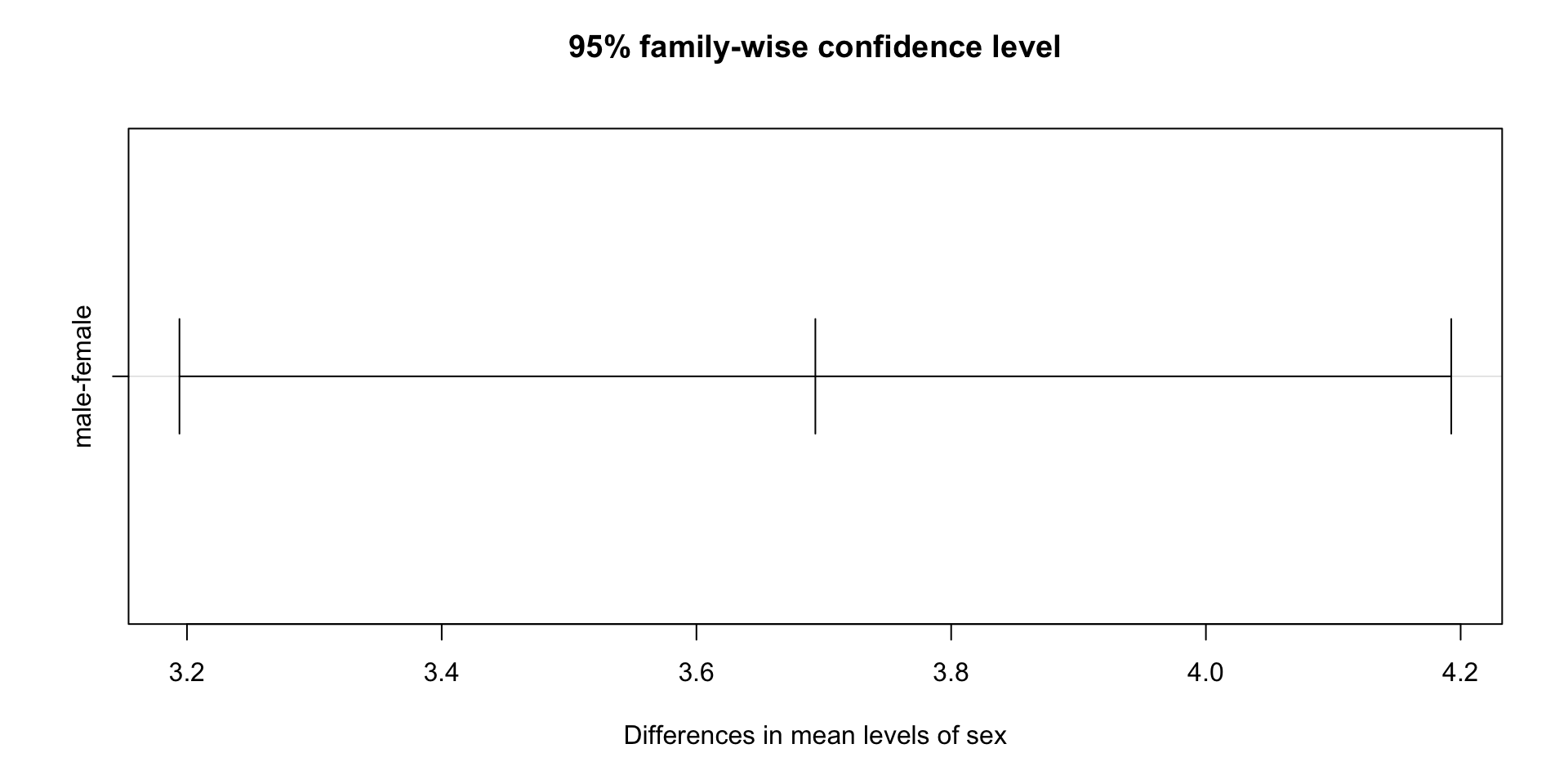
We can check how different the means of the three groups are:
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = bp ~ group, data = gather(BP, key = "group", value = "bp"))
$group
diff lwr upr p adj
old-middle 5 -0.09748151 10.09748151 0.0543408
young-middle -5 -10.09748151 0.09748151 0.0543408
young-old -10 -15.09748151 -4.90251849 0.0010202- We can also visualize the difference:
4 Example
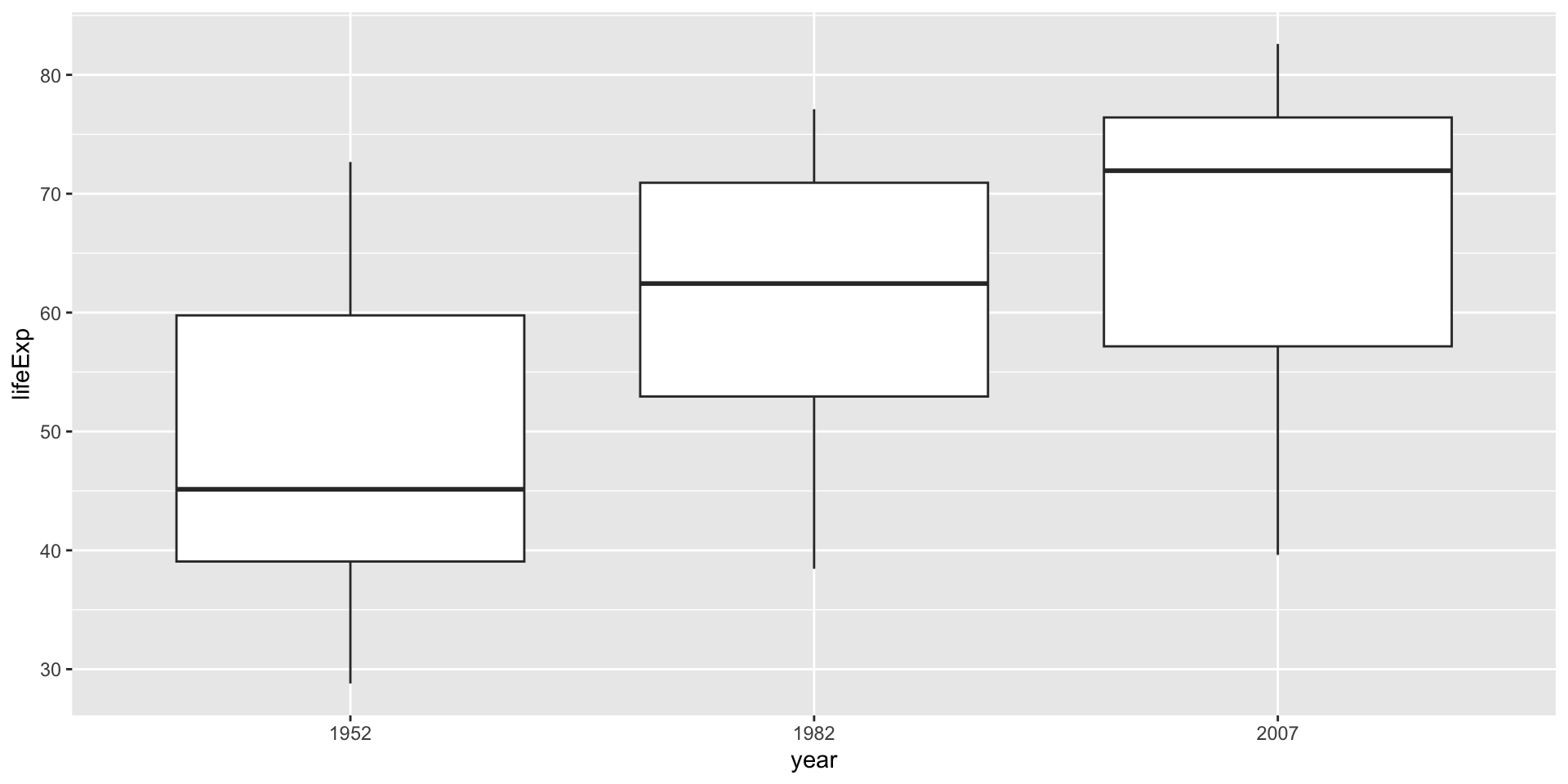
We used a very simple example to show how to calculate f-value. Now, let’s use a real data set to do the same thing. We use
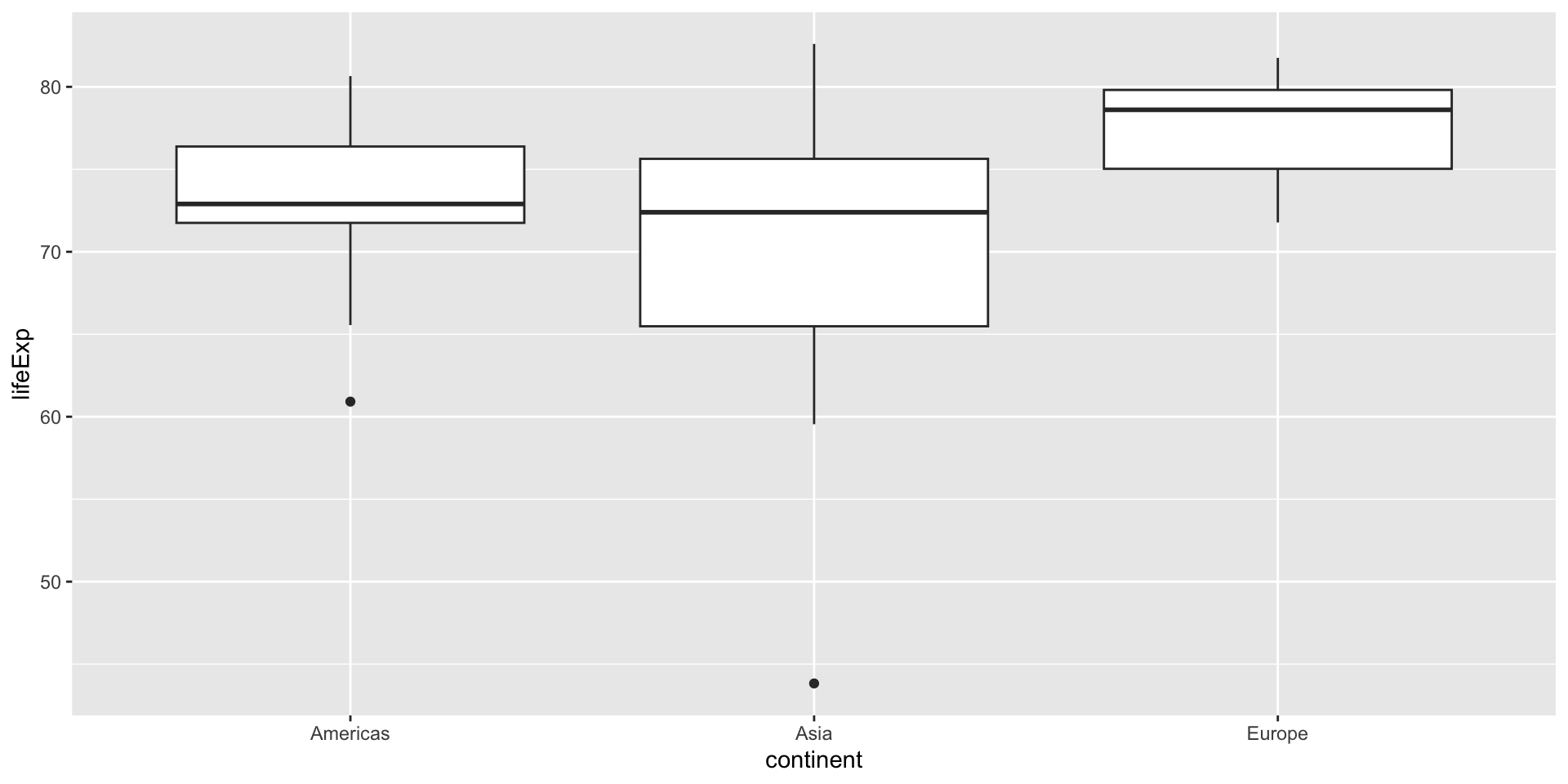
gapminderdata set to compare the life expectancy of three different continents: Asia, Europe, and Americas.First, let’s visualize the data:
-
According to the boxplot, we make our hypothesis:
H0: The mean life expectancy is the same across the three continents.
H1: The mean life expectancy differs at least for one continent among the three.
We use R to do the test:
Df Sum Sq Mean Sq F value Pr(>F)
continent 2 755.6 377.8 11.63 3.42e-05 ***
Residuals 85 2760.3 32.5
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Based on the result, we can see that the p-value is close to 0, so we reject the null hypothesis and conclude that there is a significant difference in life expectancy among the three continents.
Which and how different?
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = lifeExp ~ continent, data = filter(gapminder, year == 2007, continent %in% c("Asia", "Europe", "Americas")))
$continent
diff lwr upr p adj
Asia-Americas -2.879635 -6.4839802 0.7247099 0.1432634
Europe-Americas 4.040480 0.3592746 7.7216854 0.0279460
Europe-Asia 6.920115 3.4909215 10.3493088 0.0000189- As we can see, the main difference is between Europe and the other two continents, which is the same as we can see from the boxplot.
5 Two-Way ANOVA
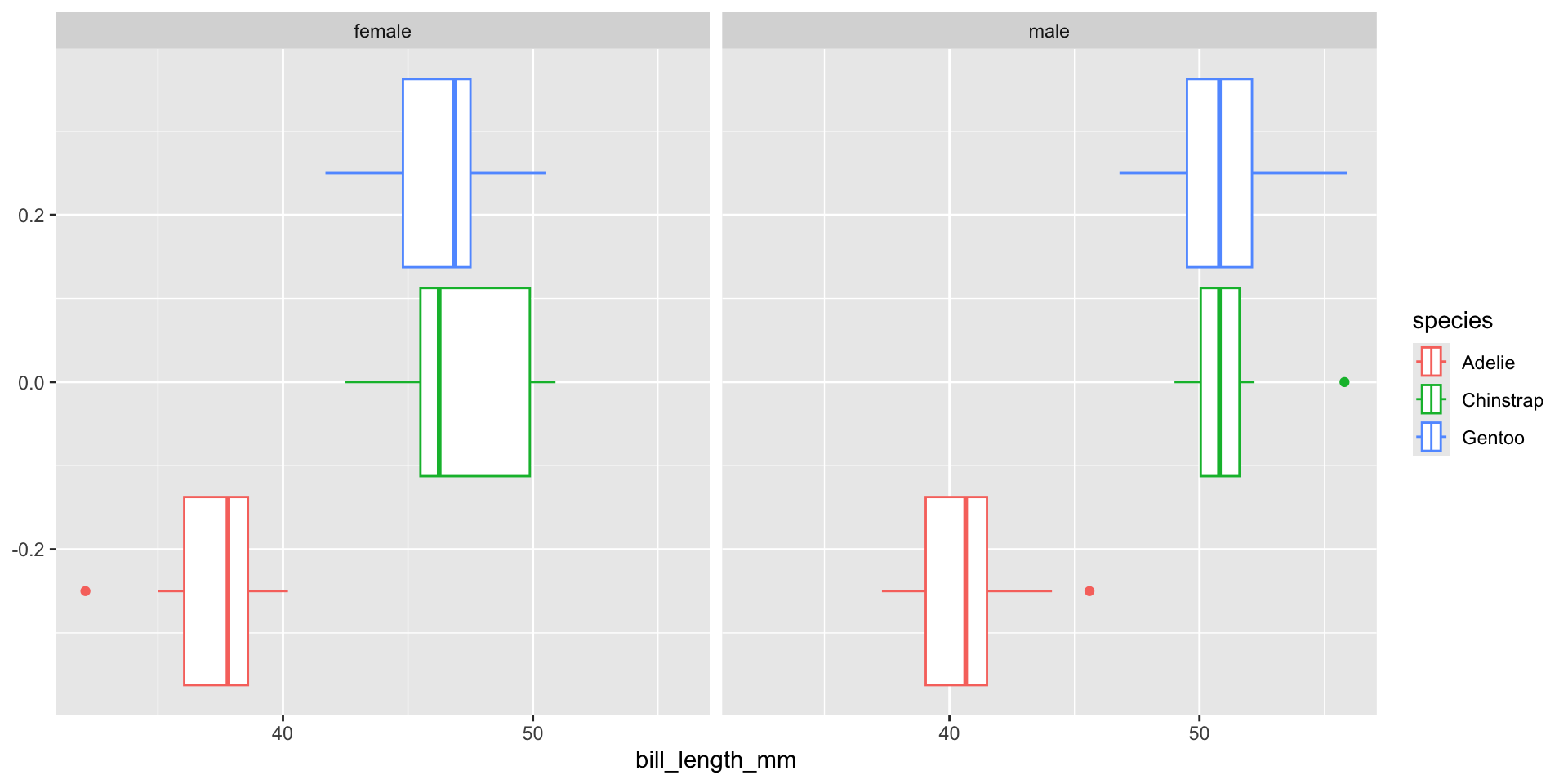
- In the above example, we only consider the ages which divided by three groups. If we also consider the gender, then we need to use two-way ANOVA.
Seen from the plots, in two-way ANOVA, we not only compare among species, also compare between sexes.
-
Research questions
Is there a significant difference in bill length across different species?
Do male and female penguins exhibit variations in bill length?
Is there a statistically significant interaction effect between species and sex concerning bill length?
-
Hypothese
-
Research Question 1: Is there a significant difference in bill length across different species?
- Null Hypothesis (H0): The mean bill length is the same across all species.
- Alternative Hypothesis (H1): There is a significant difference in the mean bill length among different species.
-
Research Question 2: Do male and female individuals exhibit variations in bill length?
Null Hypothesis (H0): There is no difference in the mean bill length between male and female individuals.
Alternative Hypothesis (H1): There is a significant difference in the mean bill length between male and female individuals.
-
Research Question 3: Is there a statistically significant interaction effect between species and sex concerning bill length?
Null Hypothesis (H0): There is no interaction effect between species and sex on bill length.
Alternative Hypothesis (H1): There is a statistically significant interaction effect between species and sex concerning bill length.
-
We use R to do the test:
Df Sum Sq Mean Sq F value Pr(>F)
species 2 2778.3 1389.2 299.861 <2e-16 ***
sex 1 435.4 435.4 93.979 <2e-16 ***
species:sex 2 12.8 6.4 1.382 0.255
Residuals 111 514.2 4.6
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Based on the result, we can see that the p-value is close to 0, so we reject the null hypothesis and conclude that there is a significant difference in bill length among the three species.
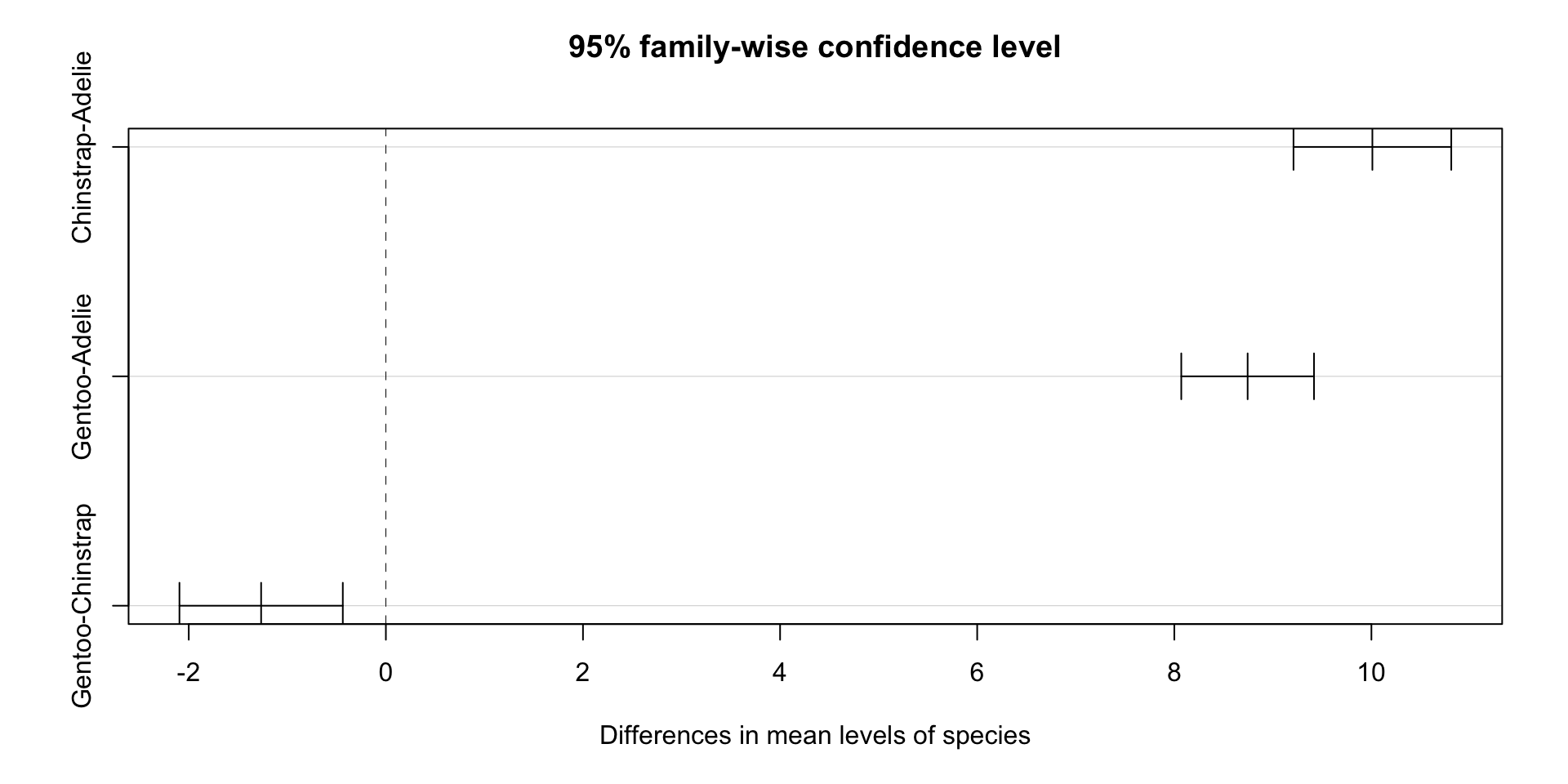
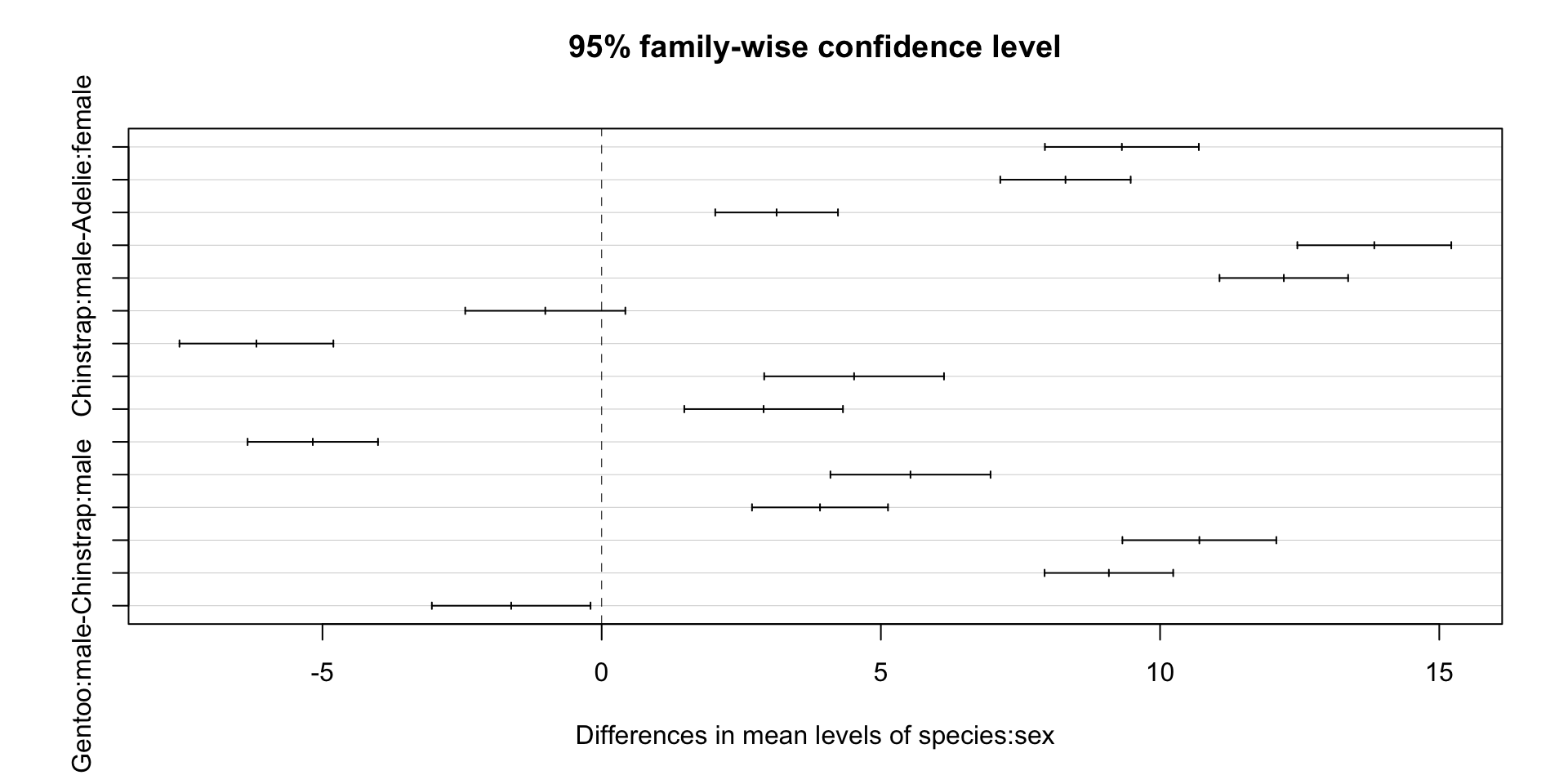
Which and how different?
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = bill_length_mm ~ species * sex, data = drop_na(penguins, sex))
$species
diff lwr upr p adj
Chinstrap-Adelie 10.009851 9.209424 10.8102782 0.0000000
Gentoo-Adelie 8.744095 8.070779 9.4174106 0.0000000
Gentoo-Chinstrap -1.265756 -2.094535 -0.4369773 0.0010847
$sex
diff lwr upr p adj
male-female 3.693368 3.194089 4.192646 0
$`species:sex`
diff lwr upr p adj
Chinstrap:female-Adelie:female 9.315995 7.937732 10.6942588 0.0000000
Gentoo:female-Adelie:female 8.306259 7.138639 9.4738788 0.0000000
Adelie:male-Adelie:female 3.132877 2.034137 4.2316165 0.0000000
Chinstrap:male-Adelie:female 13.836583 12.458320 15.2148470 0.0000000
Gentoo:male-Adelie:female 12.216236 11.064727 13.3677453 0.0000000
Gentoo:female-Chinstrap:female -1.009736 -2.443514 0.4240412 0.3338130
Adelie:male-Chinstrap:female -6.183118 -7.561382 -4.8048548 0.0000000
Chinstrap:male-Chinstrap:female 4.520588 2.910622 6.1305547 0.0000000
Gentoo:male-Chinstrap:female 2.900241 1.479553 4.3209291 0.0000002
Adelie:male-Gentoo:female -5.173382 -6.341002 -4.0057622 0.0000000
Chinstrap:male-Gentoo:female 5.530325 4.096547 6.9641020 0.0000000
Gentoo:male-Gentoo:female 3.909977 2.692570 5.1273846 0.0000000
Chinstrap:male-Adelie:male 10.703707 9.325443 12.0819703 0.0000000
Gentoo:male-Adelie:male 9.083360 7.931851 10.2348686 0.0000000
Gentoo:male-Chinstrap:male -1.620347 -3.041035 -0.1996591 0.0149963As we can see, except one interact is not significant, all other within and interacts are significant.
Plot
6 Repeated Measures ANOVA (Paired ANOVA)
- Repeated measures ANOVA analyses the effects of one or more within-subjects (repeated measures) factors on a dependent variable. It’s used when the same subjects are measured multiple times under different conditions. It’s like a paired t-test, but for more than two conditions.
- Now we use R to do the test:
Error: country
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 141 52225 370.4
Error: country:year
Df Sum Sq Mean Sq F value Pr(>F)
year 2 24036 12018 588.4 <2e-16 ***
Residuals 282 5760 20
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Based on the result, we can see that the p-value is close to 0, so we reject the null hypothesis and conclude that there is a significant difference in life expectancy among the three years.
7 Assumptions
-
ANOVA has assumptions that need to be met, including the normality of data, homogeneity of variances, and independence of observations. Violation of these assumptions might affect the reliability of the ANOVA results.
Normality: The dependent variable should be approximately normally distributed for each group of the independent variable.
Homogeneity of variances: The variance of the dependent variable should be approximately equal for each group of the independent variable.
Independence: The observations should be independent of each other.
We can use R to check the assumptions. If the assumptions are not met, we can use non-parametric tests, such as Kruskal-Wallis test, to do the test.
8 Recap
-
In this lesson, we learned:
One-way ANOVA
Two-way ANOVA
Repeated measures ANOVA
In next lesson, we will learn how to do regression analysis in R.
Thank you!