Introduction to Survival Analysis in R
生存模型
1 Setup
2 Introduction
3 Kaplan-Meier estimator of the survival function
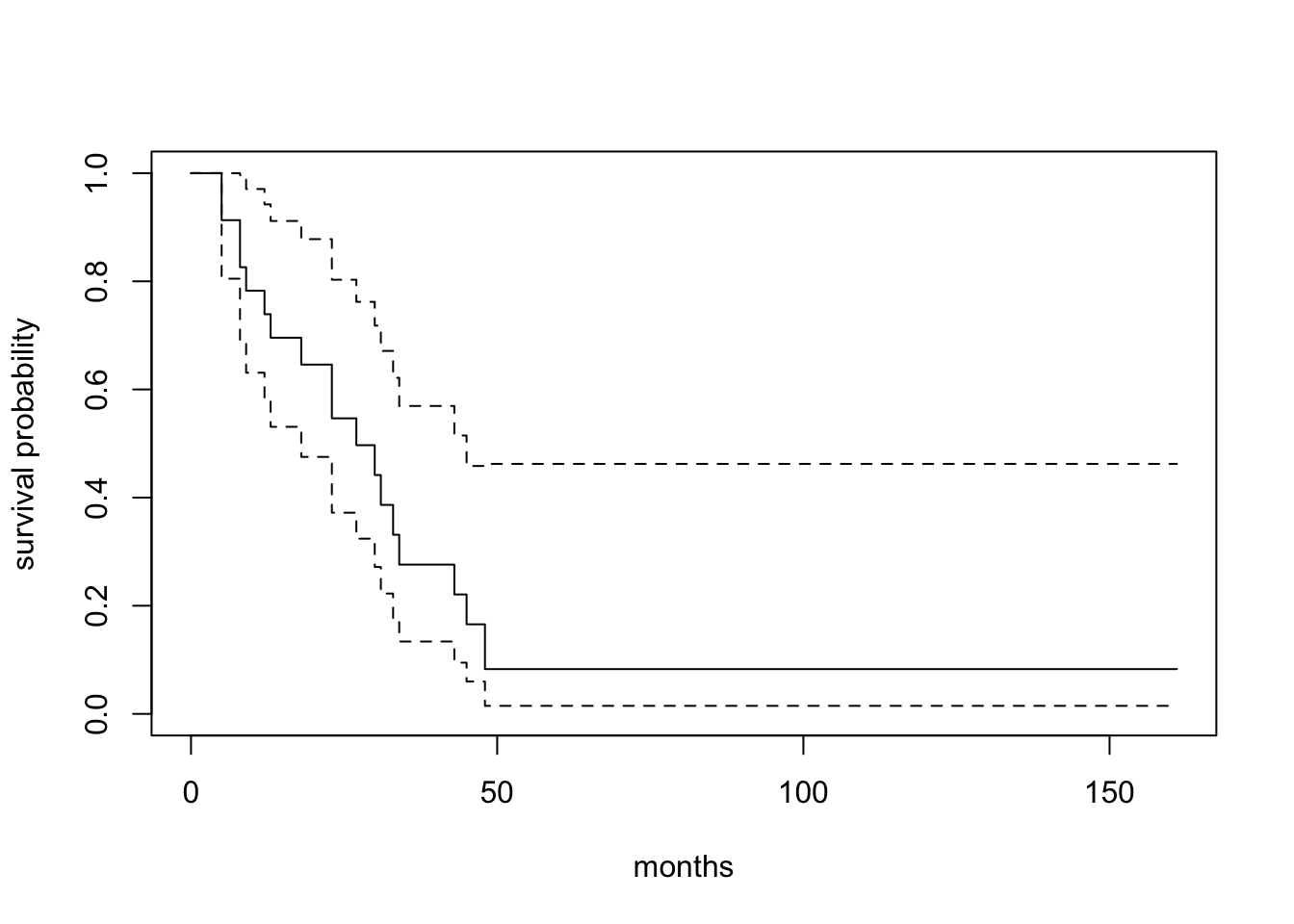
3.1 Kaplan-Meier estimation with survfit()
Call: survfit(formula = Surv(time, status) ~ 1, data = aml)
n events median 0.95LCL 0.95UCL
[1,] 23 18 27 18 45# A tibble: 6 × 8
time n.risk n.event n.censor estimate std.error conf.high conf.low
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 5 23 2 0 0.913 0.0643 1 0.805
2 8 21 2 0 0.826 0.0957 0.996 0.685
3 9 19 1 0 0.783 0.110 0.971 0.631
4 12 18 1 0 0.739 0.124 0.942 0.580
5 13 17 1 1 0.696 0.138 0.912 0.531
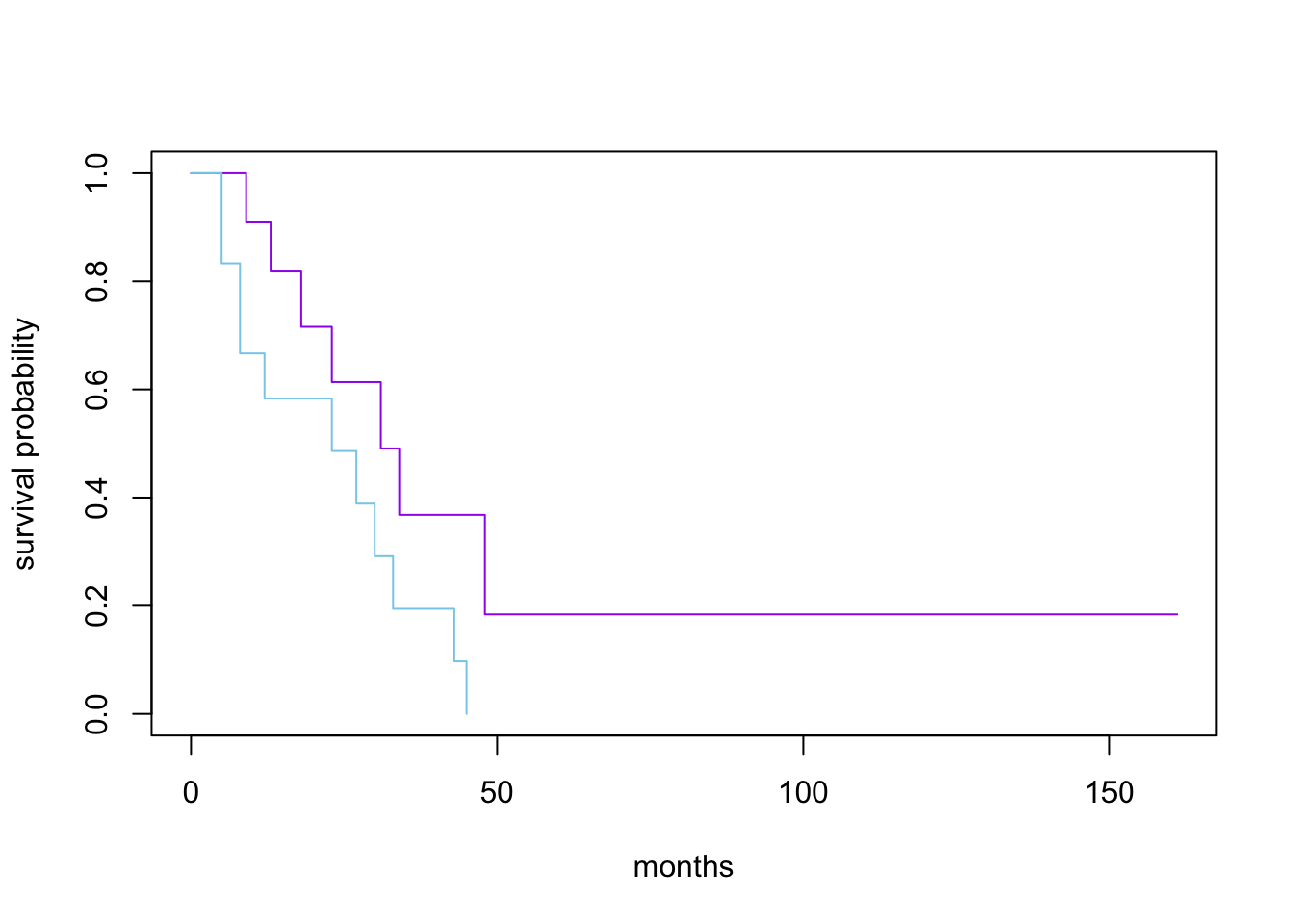
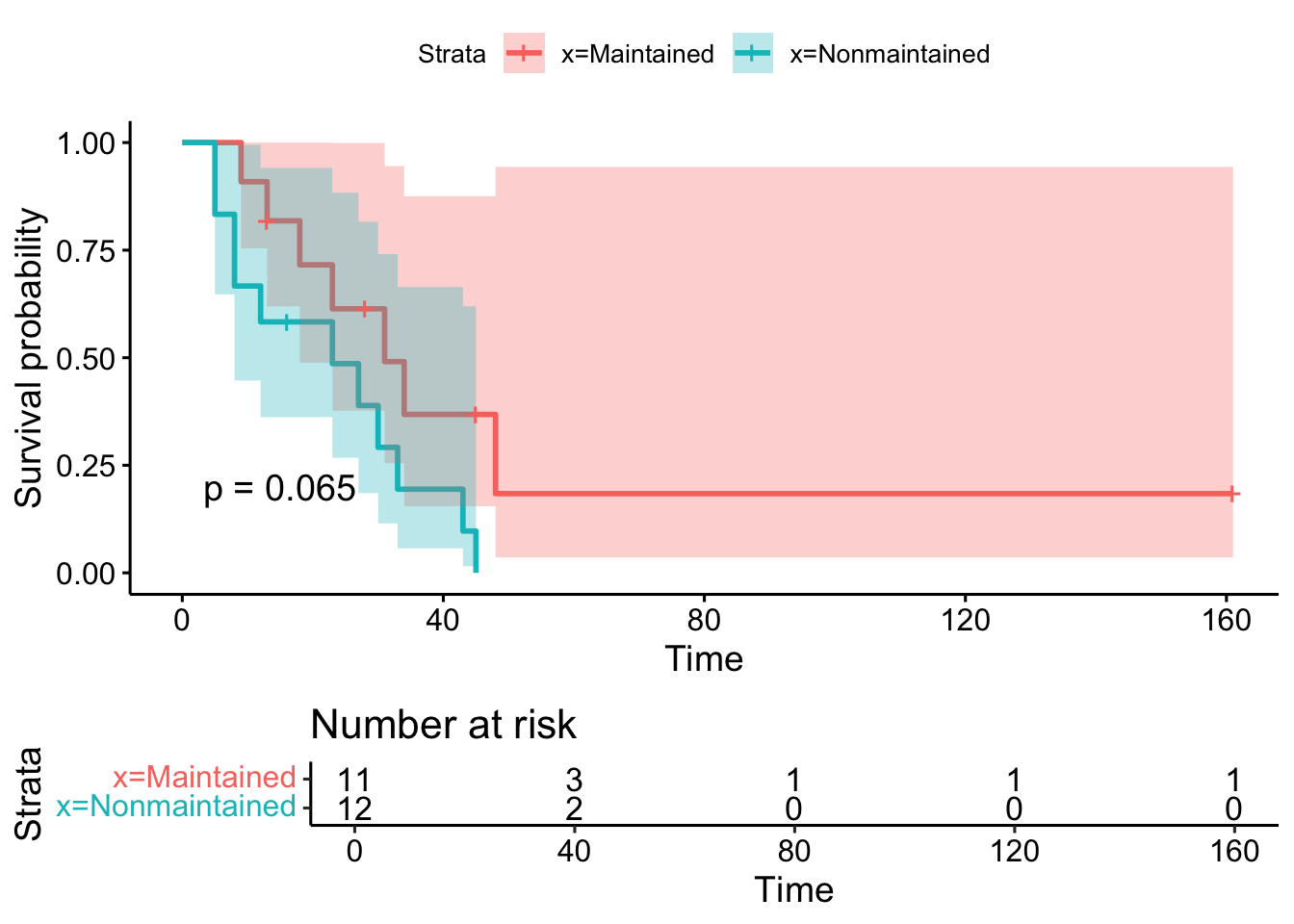
6 16 15 0 1 0.696 0.138 0.912 0.5313.2 Stratified Kaplan-Meier estimates
Call: survfit(formula = Surv(time, status) ~ x, data = aml)
n events median 0.95LCL 0.95UCL
x=Maintained 11 7 31 18 NA
x=Nonmaintained 12 11 23 8 NA# A tibble: 6 × 9
time n.risk n.event n.censor estimate std.error conf.high conf.low strata
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 9 11 1 0 0.909 0.0953 1 0.754 x=Maintai…
2 13 10 1 1 0.818 0.142 1 0.619 x=Maintai…
3 18 8 1 0 0.716 0.195 1 0.488 x=Maintai…
4 23 7 1 0 0.614 0.249 0.999 0.377 x=Maintai…
5 28 6 0 1 0.614 0.249 0.999 0.377 x=Maintai…
6 31 5 1 0 0.491 0.334 0.946 0.255 x=Maintai…4 Comparing survival functions with survdiff()
We often want to test the hypothesis:
H0: survival curves across 2 or more groups are equivalent HA: survival curves across 2 or more groups are not equivalent
The log-rank statistic is one popular method to evaluate this hypothesis.
Under the null, the log-rank statistic is χ2 distributed with g−1 degrees of freedom.
The function survdiff() performs the log-rank test by default.
Call:
survdiff(formula = Surv(time, status) ~ x, data = aml)
N Observed Expected (O-E)^2/E (O-E)^2/V
x=Maintained 11 7 10.69 1.27 3.4
x=Nonmaintained 12 11 7.31 1.86 3.4
Chisq= 3.4 on 1 degrees of freedom, p= 0.07 Code
Call:
survdiff(formula = Surv(time, status) ~ x, data = aml, rho = 1)
N Observed Expected (O-E)^2/E (O-E)^2/V
x=Maintained 11 3.85 6.14 0.859 2.78
x=Nonmaintained 12 7.18 4.88 1.081 2.78
Chisq= 2.8 on 1 degrees of freedom, p= 0.1 5 Exercise 1
The veteran data set describes survival times for veterans with lung cancer.
Variables:
time: survival time status: status, 0=censored, 1=dead trt: treatment, 1=standard, 2=test
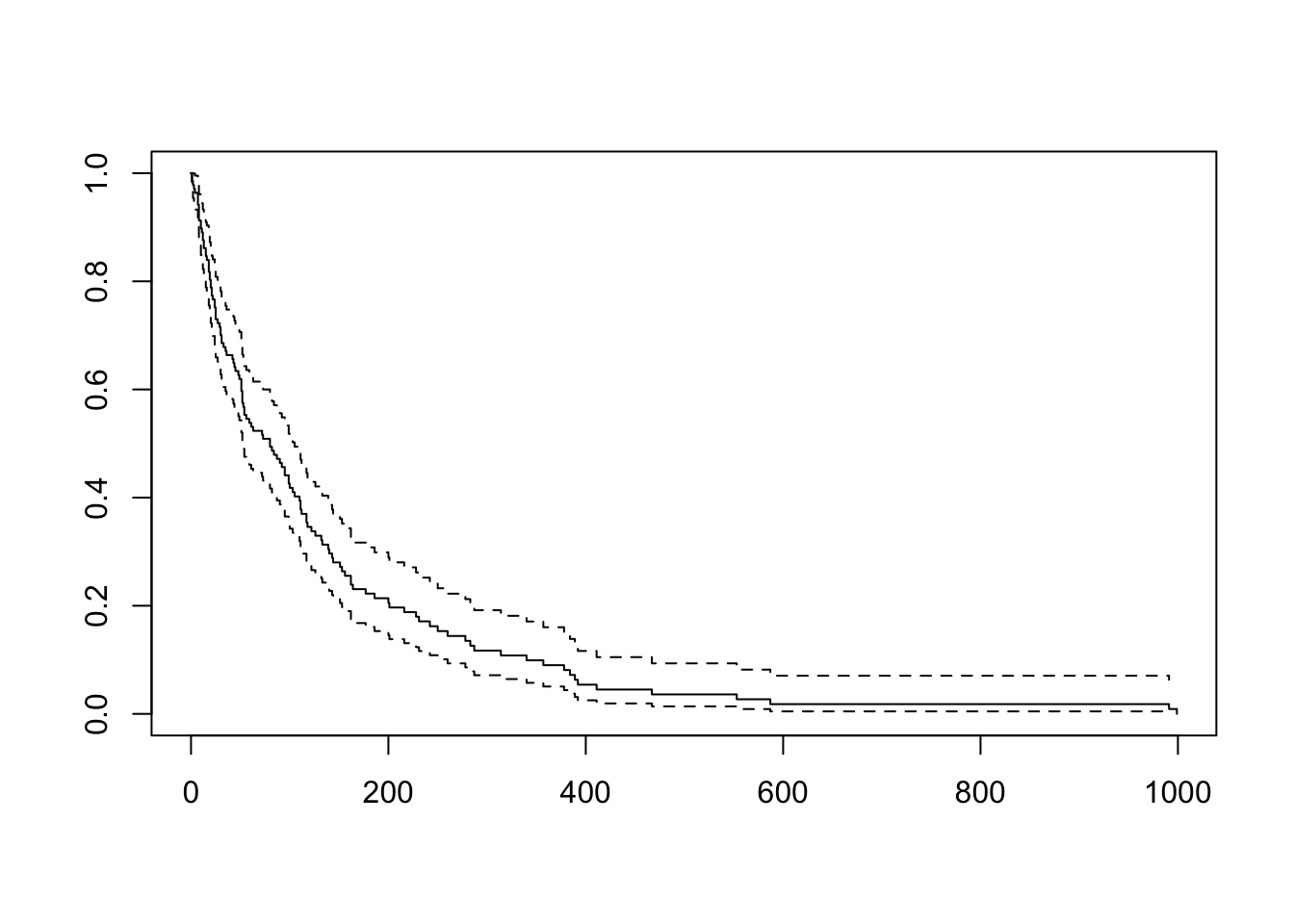
- Create a graph and table of the Kaplan-Meier estimated survival function for the entire data set. What is the median survival time?
Call: survfit(formula = Surv(time, status) ~ 1, data = veteran)
n events median 0.95LCL 0.95UCL
[1,] 137 128 80 52 105# A tibble: 6 × 8
time n.risk n.event n.censor estimate std.error conf.high conf.low
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 137 2 0 0.985 0.0104 1 0.966
2 2 135 1 0 0.978 0.0128 1 0.954
3 3 134 1 0 0.971 0.0148 0.999 0.943
4 4 133 1 0 0.964 0.0166 0.995 0.933
5 7 132 3 0 0.942 0.0213 0.982 0.903
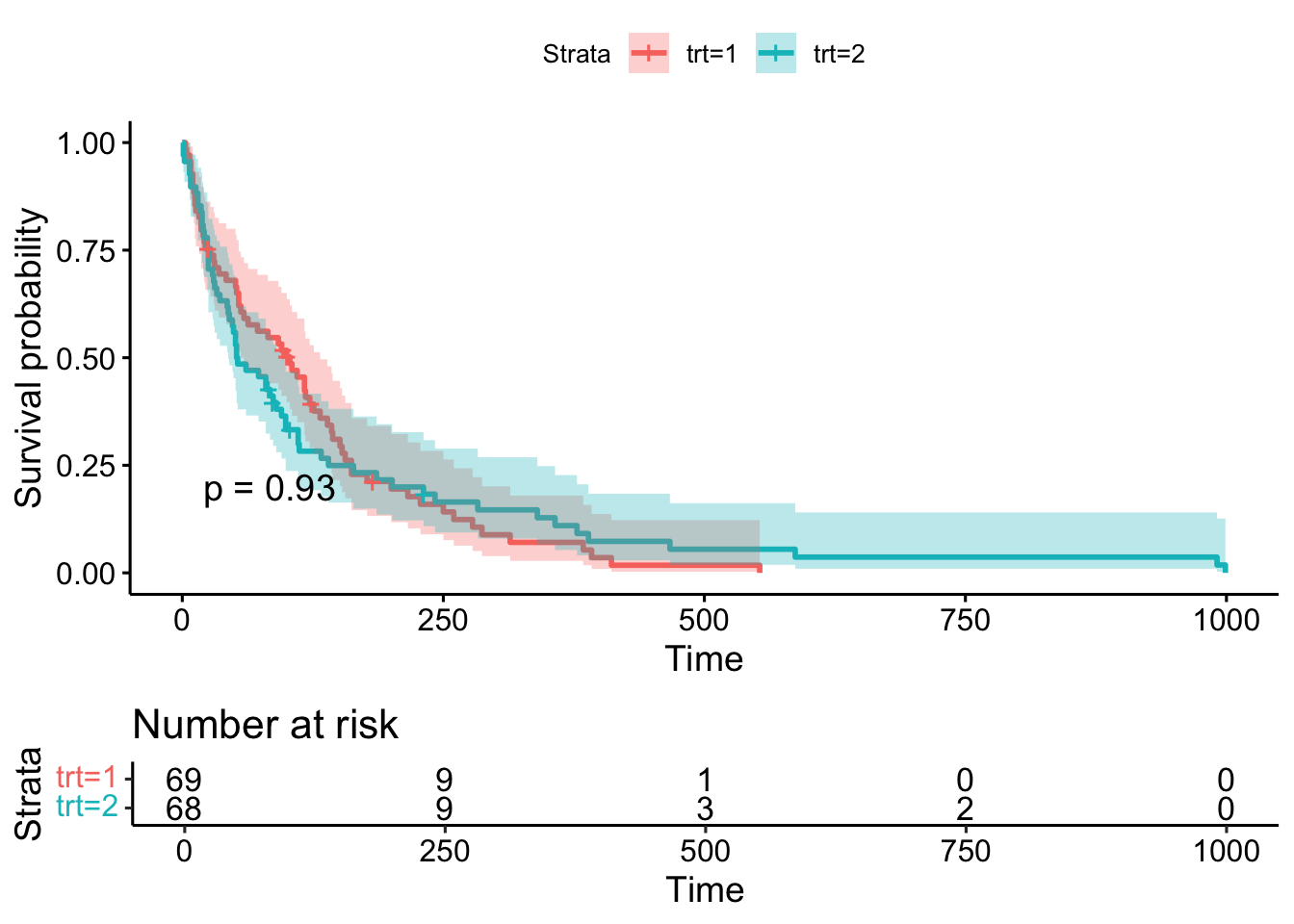
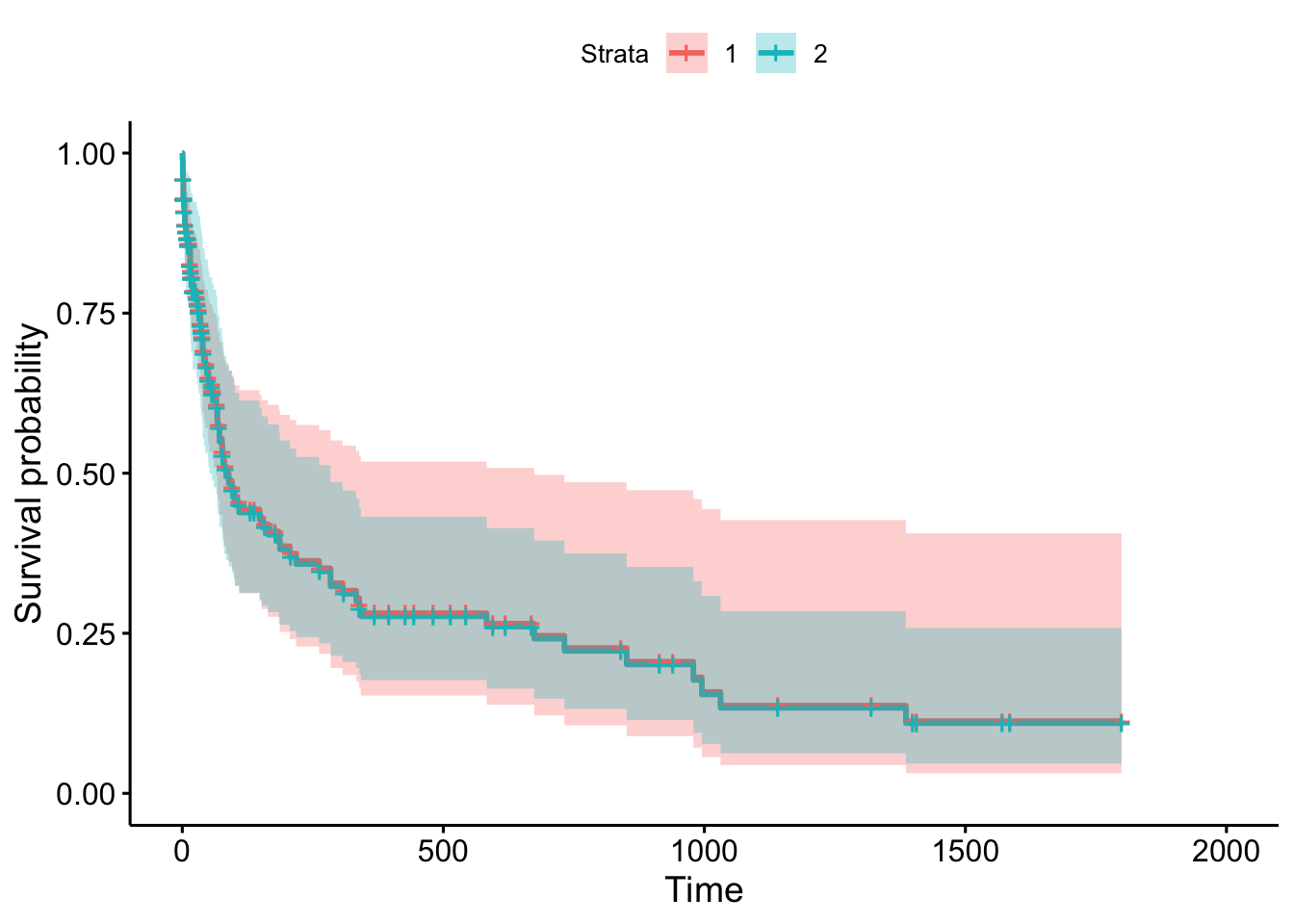
6 8 129 4 0 0.912 0.0265 0.961 0.866- Create a graph of KM estimated survival functions stratified by treatment, adding 95% confidence intervals and coloring the 2 functions. Does survival appear different for the 2 treatment groups?
Call: survfit(formula = Surv(time, status) ~ trt, data = veteran)
n events median 0.95LCL 0.95UCL
trt=1 69 64 103.0 59 132
trt=2 68 64 52.5 44 95- Use the log-rank test to provide more evidence for your assessment of survival of the 2 groups.
6 The Cox proportional hazards model
6.1 Background
6.2 The Cox proportional hazards model
6.3 Proportional hazards
Fitting a Cox model
Call:
coxph(formula = Surv(time, status) ~ age + sex + wt.loss, data = lung)
n= 214, number of events= 152
(14 observations deleted due to missingness)
coef exp(coef) se(coef) z Pr(>|z|)
age 0.0200882 1.0202913 0.0096644 2.079 0.0377 *
sex -0.5210319 0.5939074 0.1743541 -2.988 0.0028 **
wt.loss 0.0007596 1.0007599 0.0061934 0.123 0.9024
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
age 1.0203 0.9801 1.0011 1.0398
sex 0.5939 1.6838 0.4220 0.8359
wt.loss 1.0008 0.9992 0.9887 1.0130
Concordance= 0.612 (se = 0.027 )
Likelihood ratio test= 14.67 on 3 df, p=0.002
Wald test = 13.98 on 3 df, p=0.003
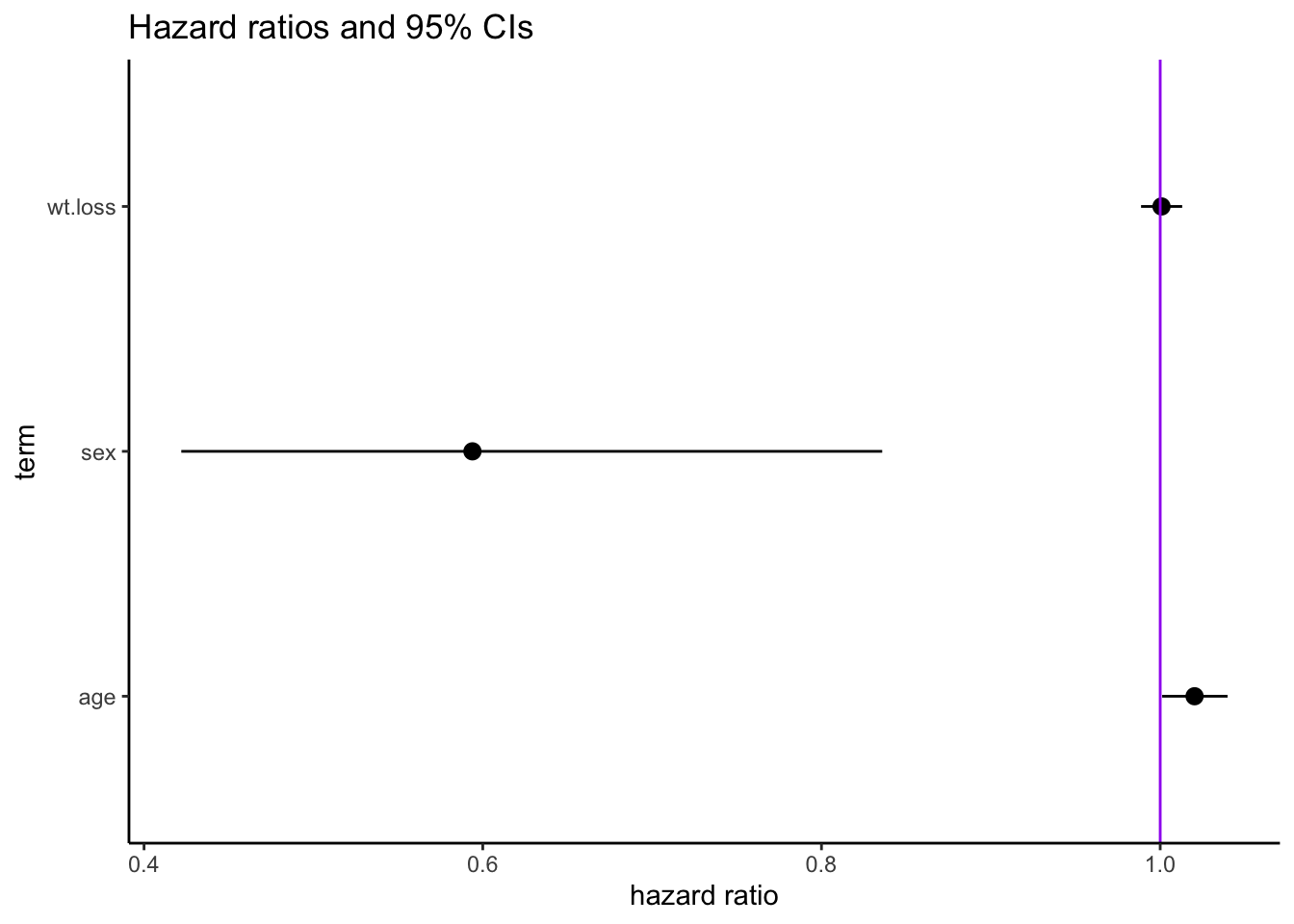
Score (logrank) test = 14.24 on 3 df, p=0.0036.4 Tidy coxph() results
# A tibble: 3 × 7
term estimate std.error statistic p.value conf.low conf.high
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 age 1.02 0.00966 2.08 0.0377 1.00 1.04
2 sex 0.594 0.174 -2.99 0.00280 0.422 0.836
3 wt.loss 1.00 0.00619 0.123 0.902 0.989 1.01 Code
lung.cox.tab |>
ggplot(aes(
x = estimate,
y = term,
xmin = conf.low,
xmax = conf.high
)) +
geom_pointrange() + # plots center point (x) and range (xmin, xmax)
geom_vline(xintercept = 1, color = "purple") + # vertical line at HR=1
labs(x = "hazard ratio", title = "Hazard ratios and 95% CIs") +
theme_classic()6.5 Predicting survival with Cox estimates
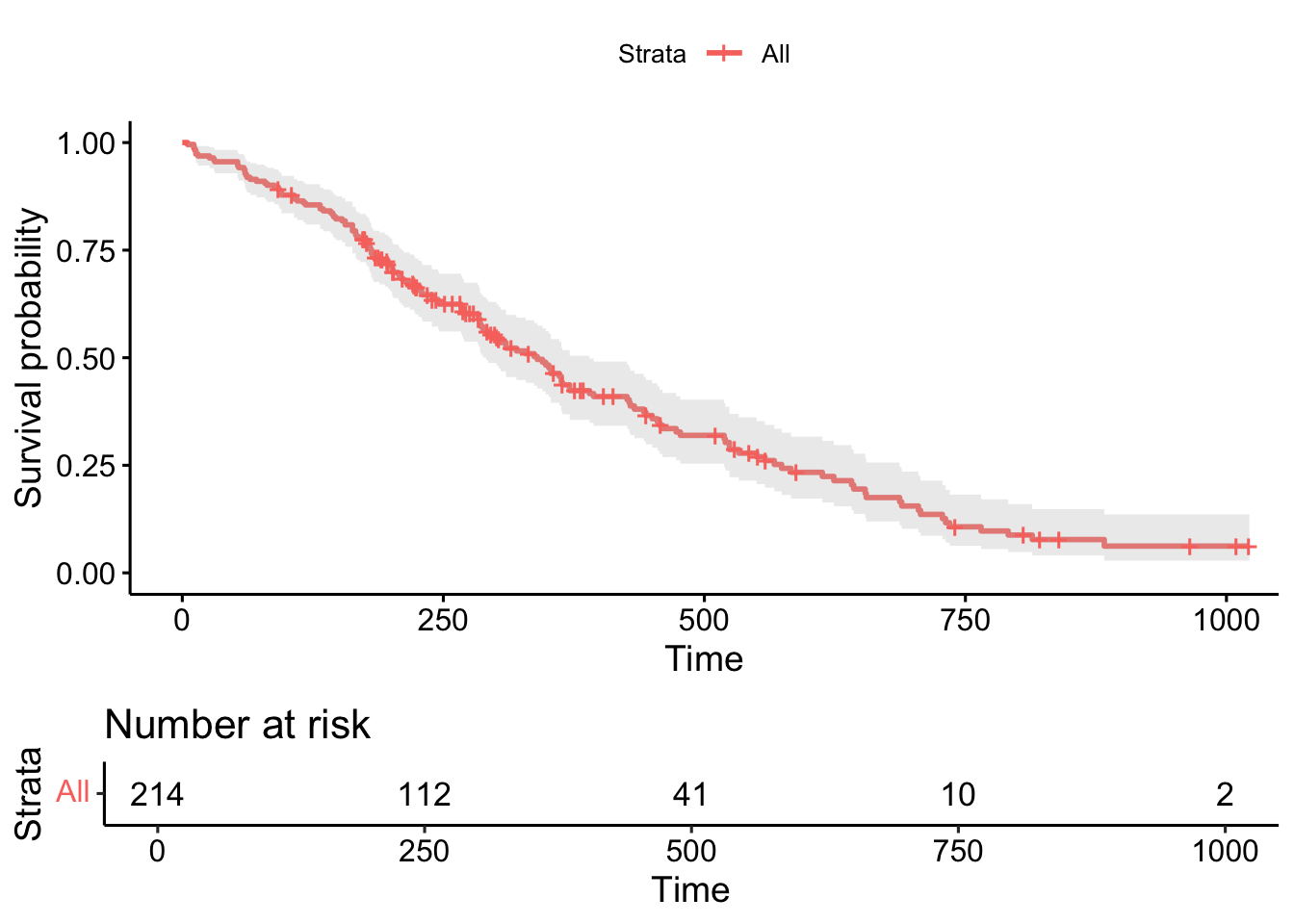
Code
# A tibble: 179 × 8
time n.risk n.event n.censor estimate std.error conf.high conf.low
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 5 214 1 0 0.996 0.00443 1 0.987
2 11 213 2 0 0.987 0.00772 1 0.972
3 12 211 1 0 0.982 0.00894 1.00 0.965
4 13 210 2 0 0.973 0.0110 0.995 0.953
5 15 208 1 0 0.969 0.0120 0.992 0.946
6 26 207 1 0 0.964 0.0128 0.989 0.940
7 30 206 1 0 0.960 0.0137 0.986 0.935
8 31 205 1 0 0.955 0.0145 0.983 0.929
9 53 204 2 0 0.946 0.0160 0.976 0.917
10 54 202 1 0 0.942 0.0167 0.973 0.912
# ℹ 169 more rowsCode
age sex wt.loss
1 62.44737 1 9.831776
2 62.44737 2 9.831776Code
# A tibble: 179 × 12
time n.risk n.event n.censor estimate.1 estimate.2 std.error.1 std.error.2
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 5 214 1 0 0.995 0.997 0.00546 0.00327
2 11 213 2 0 0.984 0.990 0.00953 0.00577
3 12 211 1 0 0.978 0.987 0.0111 0.00674
4 13 210 2 0 0.967 0.980 0.0137 0.00844
5 15 208 1 0 0.962 0.977 0.0148 0.00921
6 26 207 1 0 0.956 0.974 0.0159 0.00995
7 30 206 1 0 0.951 0.971 0.0170 0.0107
8 31 205 1 0 0.945 0.967 0.0180 0.0113
9 53 204 2 0 0.934 0.960 0.0199 0.0127
10 54 202 1 0 0.929 0.957 0.0208 0.0133
# ℹ 169 more rows
# ℹ 4 more variables: conf.high.1 <dbl>, conf.high.2 <dbl>, conf.low.1 <dbl>,
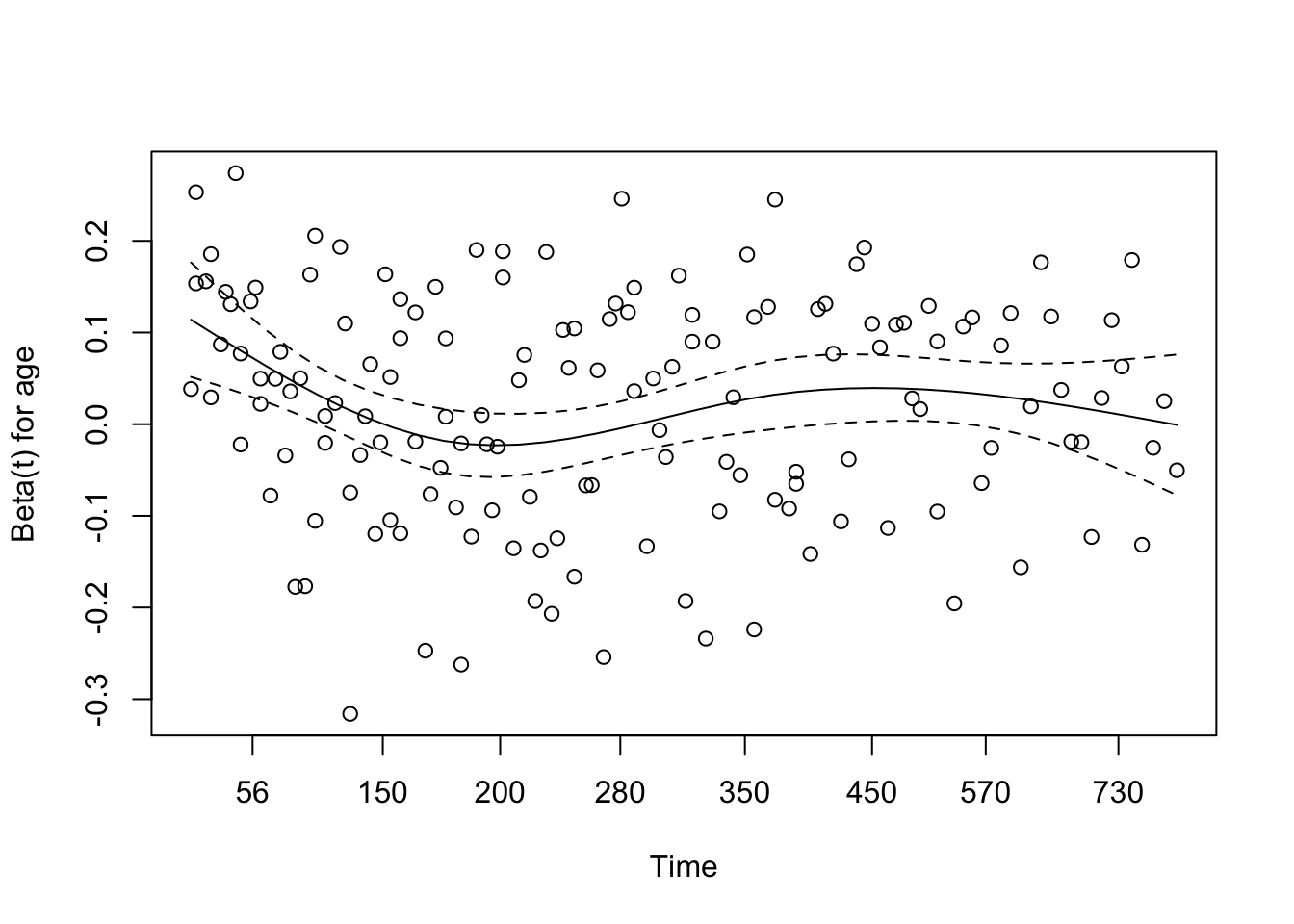
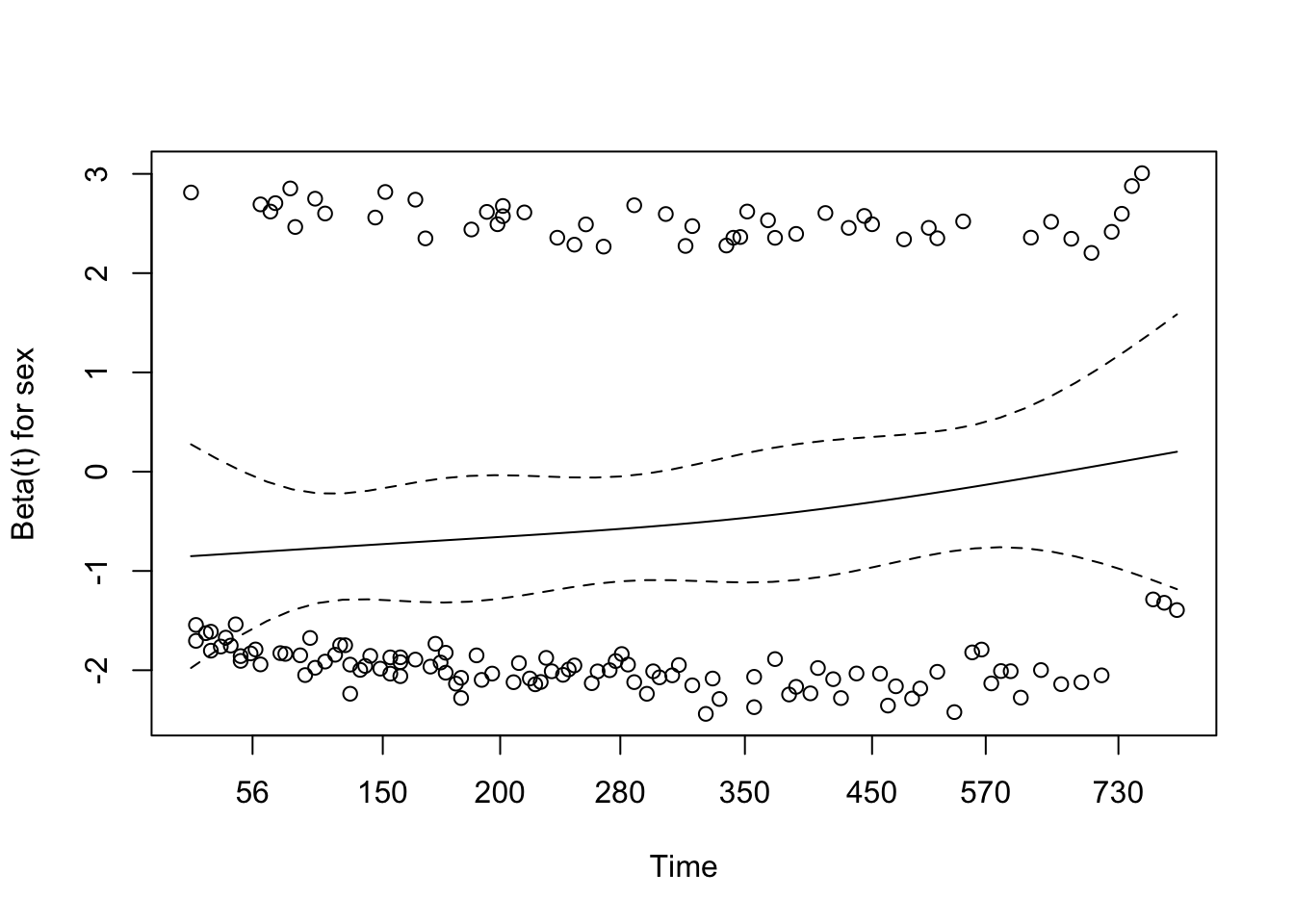
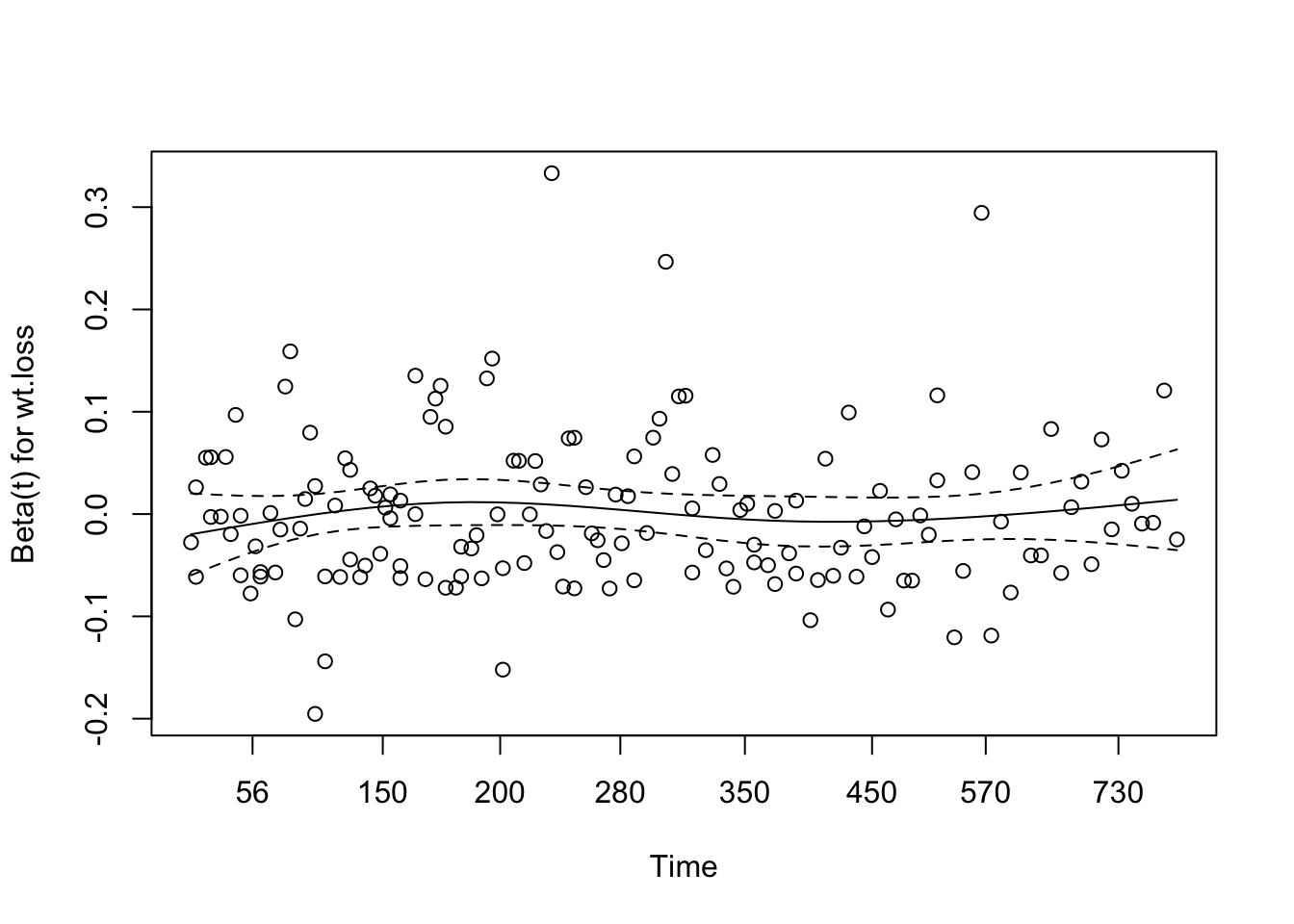
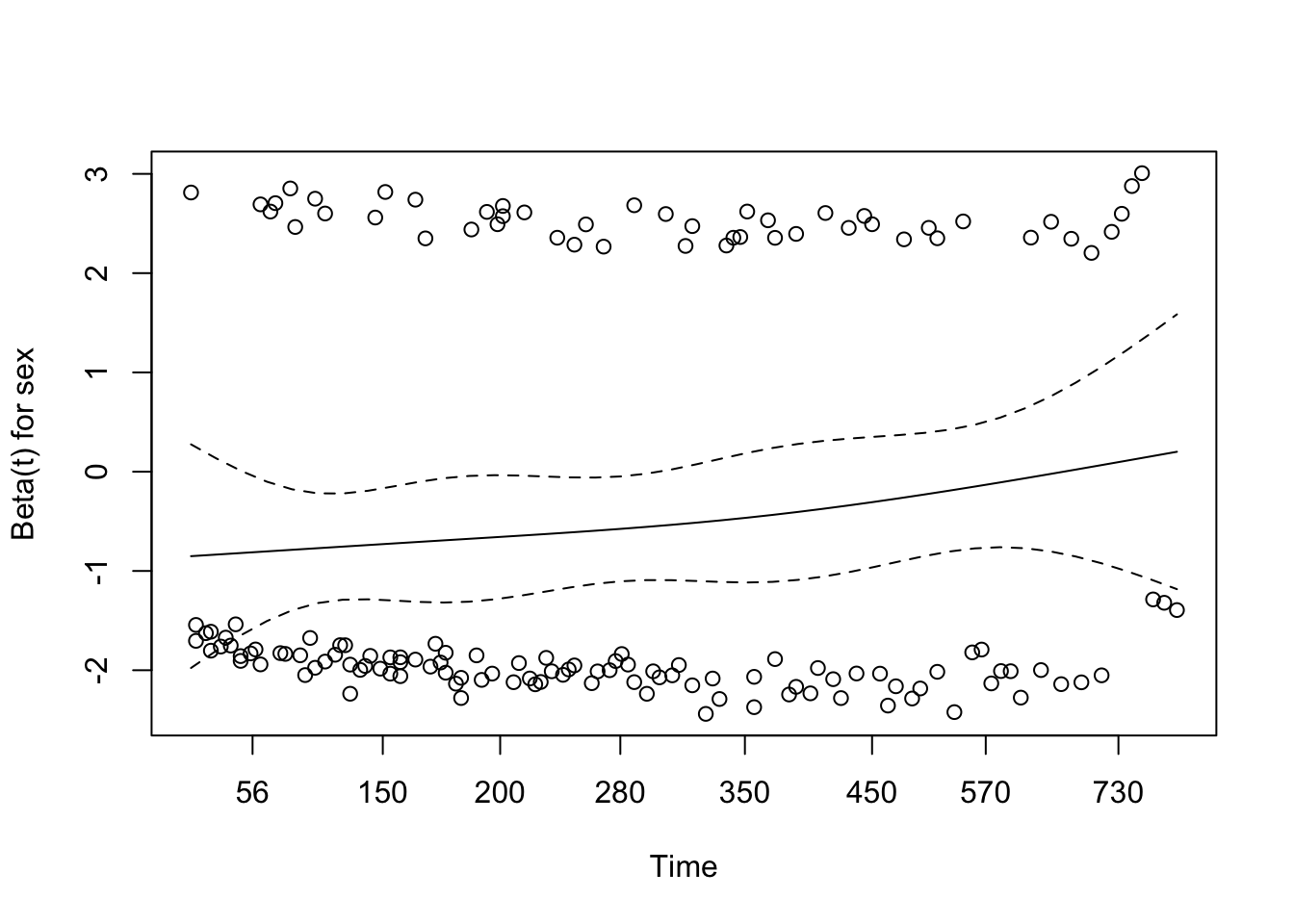
# conf.low.2 <dbl>7 Assessing the proportional hazards assumption
A chi-square test based on Schoenfeld residuals is available with cox.zph() to test the hypothesis:
H0: covariate effect is constant (proportional) over time Ha: covariate effect changes over time
chisq df p
age 0.5077 1 0.48
sex 2.5489 1 0.11
wt.loss 0.0144 1 0.90
GLOBAL 3.0051 3 0.398 Dealing with proportional hazards violations
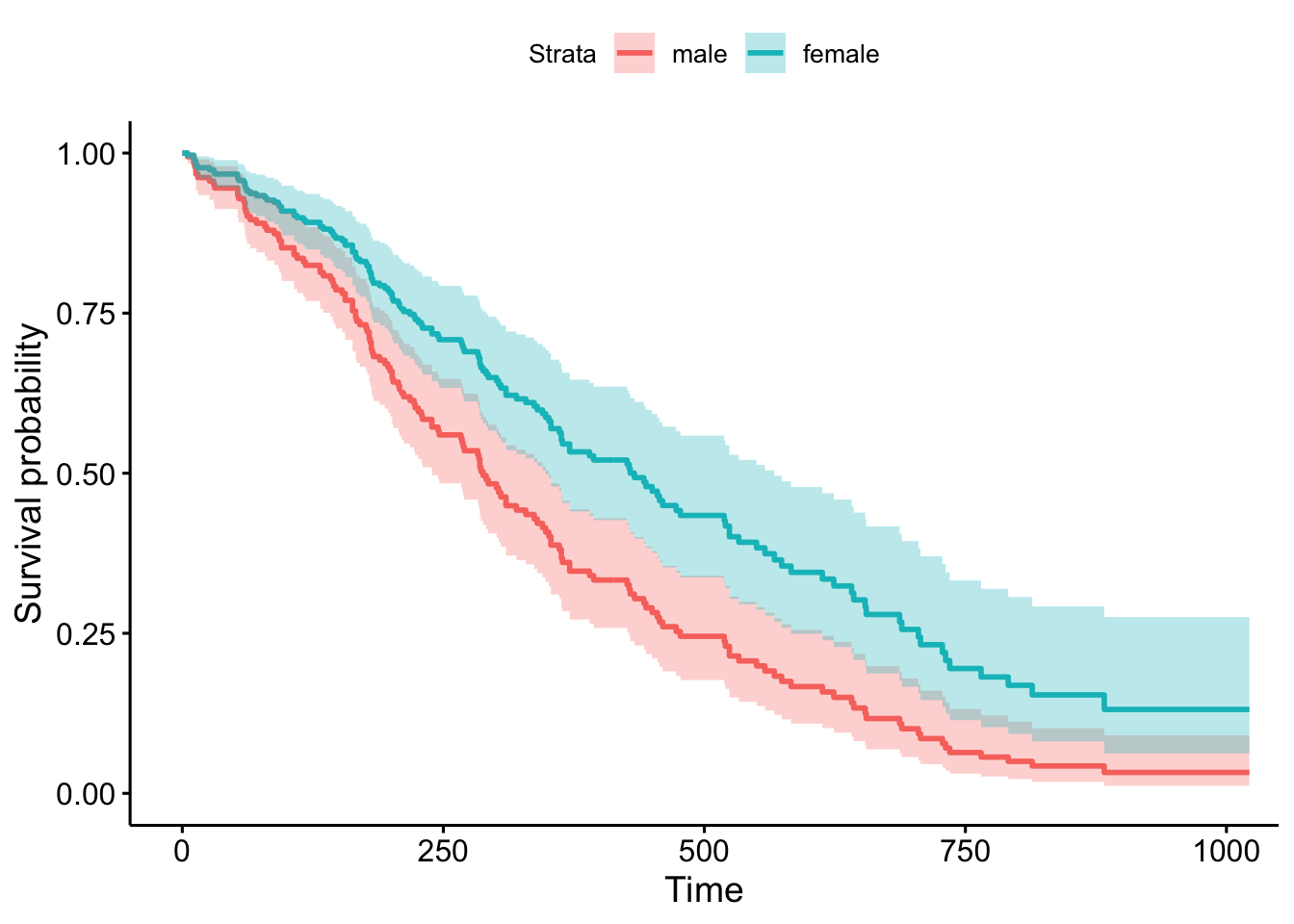
Call:
coxph(formula = Surv(time, status) ~ age + sex + wt.loss, data = lung)
n= 214, number of events= 152
(14 observations deleted due to missingness)
coef exp(coef) se(coef) z Pr(>|z|)
age 0.0200882 1.0202913 0.0096644 2.079 0.0377 *
sex -0.5210319 0.5939074 0.1743541 -2.988 0.0028 **
wt.loss 0.0007596 1.0007599 0.0061934 0.123 0.9024
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
age 1.0203 0.9801 1.0011 1.0398
sex 0.5939 1.6838 0.4220 0.8359
wt.loss 1.0008 0.9992 0.9887 1.0130
Concordance= 0.612 (se = 0.027 )
Likelihood ratio test= 14.67 on 3 df, p=0.002
Wald test = 13.98 on 3 df, p=0.003
Score (logrank) test = 14.24 on 3 df, p=0.0039 Stratified Cox model
Code
Call:
coxph(formula = Surv(time, status) ~ age + wt.loss + strata(sex),
data = lung)
n= 214, number of events= 152
(14 observations deleted due to missingness)
coef exp(coef) se(coef) z Pr(>|z|)
age 0.0192190 1.0194049 0.0096226 1.997 0.0458 *
wt.loss 0.0001412 1.0001412 0.0062509 0.023 0.9820
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
age 1.019 0.9810 1.000 1.039
wt.loss 1.000 0.9999 0.988 1.012
Concordance= 0.561 (se = 0.027 )
Likelihood ratio test= 4.09 on 2 df, p=0.1
Wald test = 3.99 on 2 df, p=0.1
Score (logrank) test = 4 on 2 df, p=0.110 Modeling time-varying coefficients
Code
Call:
coxph(formula = Surv(time, status) ~ age + wt.loss + sex + tt(sex),
data = lung, tt = function(x, t, ...) x * t)
n= 214, number of events= 152
(14 observations deleted due to missingness)
coef exp(coef) se(coef) z Pr(>|z|)
age 0.0194343 1.0196244 0.0096522 2.013 0.0441 *
wt.loss 0.0001260 1.0001261 0.0062502 0.020 0.9839
sex -0.9417444 0.3899470 0.3224791 -2.920 0.0035 **
tt(sex) 0.0013778 1.0013787 0.0008581 1.606 0.1084
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
age 1.0196 0.9808 1.0005 1.0391
wt.loss 1.0001 0.9999 0.9879 1.0125
sex 0.3899 2.5645 0.2073 0.7337
tt(sex) 1.0014 0.9986 0.9997 1.0031
Concordance= 0.613 (se = 0.027 )
Likelihood ratio test= 17.23 on 4 df, p=0.002
Wald test = 15.86 on 4 df, p=0.003
Score (logrank) test = 16.44 on 4 df, p=0.00211 Time-varying covariates
Code
Call:
coxph(formula = Surv(start, stop, event) ~ transplant + age +
surgery, data = jasa1)
n= 170, number of events= 75
coef exp(coef) se(coef) z Pr(>|z|)
transplant 0.01405 1.01415 0.30822 0.046 0.9636
age 0.03055 1.03103 0.01389 2.199 0.0279 *
surgery -0.77326 0.46150 0.35966 -2.150 0.0316 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
transplant 1.0142 0.9860 0.5543 1.8555
age 1.0310 0.9699 1.0033 1.0595
surgery 0.4615 2.1668 0.2280 0.9339
Concordance= 0.599 (se = 0.036 )
Likelihood ratio test= 10.72 on 3 df, p=0.01
Wald test = 9.68 on 3 df, p=0.02
Score (logrank) test = 10 on 3 df, p=0.02 chisq df p
transplant 0.118 1 0.73
age 0.897 1 0.34
surgery 0.097 1 0.76
GLOBAL 1.363 3 0.71Code
# plot predicted survival by transplant group at mean age and surgery=0
plotdata <- data.frame(transplant=0:1, age=-2.48, surgery=0)
surv.by.transplant <- survfit(jasa1.cox, newdata=plotdata)
ggsurvplot(surv.by.transplant, data=plotdata) # remember to supply data to ggsurvplot() for predicted survival after coxph()Code
Call:
coxph(formula = Surv(ST, State) ~ Group)
n= 12, number of events= 12
coef exp(coef) se(coef) z Pr(>|z|)
GroupA 1.2515 3.4957 0.7227 1.732 0.0833 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
GroupA 3.496 0.2861 0.848 14.41
Concordance= 0.652 (se = 0.072 )
Likelihood ratio test= 3.24 on 1 df, p=0.07
Wald test = 3 on 1 df, p=0.08
Score (logrank) test = 3.36 on 1 df, p=0.07